Group Prof. Dr. Klingler
Development of the flour beetle Tribolium
Content
- Introduction
- Short and long germ segmentation
- Appendage development
- Advancing Tribolium as model system
Introduction
We study the development of the red flour beetle, Triboliumcastaneum, because this species has many characteristics of a “typical” insect, while the classical model system Drosophila melanogaster represents an evolutionary derived state. For example, the well understood segmentation process in Drosophilaonly can work in a syncytial blastoderm where transcription factors (like Bicoid) are ableto form diffusion gradients. However, in most other insects/arthropods the majority of segments do not arise during the blastoderm stage (as in Drosophila) but from a subsequent growth process which adds one segment after the other. The formation of defined “stripes” (i.e. segmentprimordia) in a cellularized growth zone likely involves different or additional molecular mechanisms. Another major difference between Tribolium and Drosophila is the way how appendages are formed. While antennae, mouth parts and legs arise in Drosophila from imaginal discs, in Tribolium they grow out from the body wall directly, during embryogenesis – just like in most arthropods. We study these processes in Tribolium because they are of fundamental interest in themselfes, and in oder to understand the evolution of embryonic patterning.
Short and long germ segmentation
The Drosophila embryo is of the “long germ” type in that the entire larval body is setup already during the blastoderm stage. Patterning in the blastoderm is based on long-range gradient semanating from both poles of the egg. These maternal gradients provide positional information that is translated by the segmentation genecascade into segmental stripes of geneexpression. This whole process takes place in the rather static setting of the blastoderm embryo, i.e. is not influenced by embryonic growth processes – a rather atypical developmental situation.
In Tribolium, only the primordia for the anterior most six segments are formed during the blastoderm stage, andex tension of this “short” germ rudiment to the fully segmented germ band represents an entirely different pattern ingenvironment. Not only is the growth zone fully cellularized which hinders the formation of diffusion gradients. Also the successive addition of segments suggests temporal rather than spatial patterning principles. Temporal regulation by an oscillatory segmentation clock mechanism directs growth and patterning in vertebrate and lower arthropod embryos. Tribolium appears to represent an intermediate step between these segmentation mode of spiders and myriapods, and the derived long germ mode of Drosophila. Our previous work has shown that many homologs of Drosophila segmentation genes also are involved in Tribolium abdominal segmentation; current efforts are directed toward sidentifying new, short germ-specific gene functions.
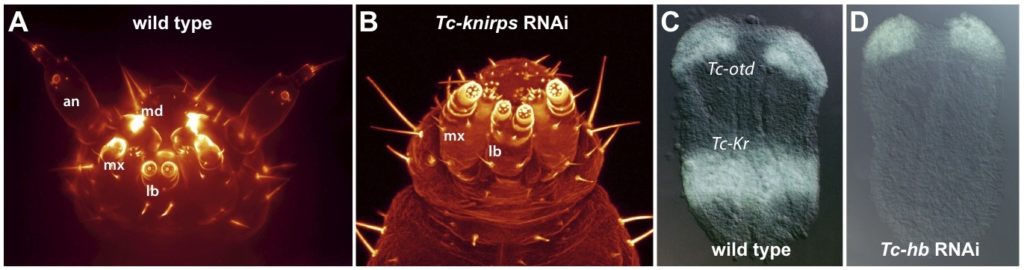
Appendage development
In Drosophila, genes functioning during imaginal disc growth have to be studied via clonalanalysis, and genome-wide screens are difficult to perform using this technology. One advantage of Tribolium is that appendage genes can be studied in the embryo, and that they can be easily identified by genome-wide RNAi screens. Understanding embryonic leg formation will provide a basis for understanding the evolution of imaginal discs, and reveal the genetic basis of arthropod limb diversity.
Improving Tribolium as model system
Together with other labs, we attempt to advance Tribolium into a system which approaches Drosophila in its suitability for the analysis of development. Recent progress includes the demonstration that systemic RNAi works in Tribolium, and that germ line transformation can be achieved with high efficiency. In 2008 the genome of Tribolium has been published, opening the way for large-scale screens and in silico analysis. A genome-wide RNAi screen aiming to identify all developmental genes in Triboliumis currently performed in Göttingen and Erlangen (Forschungsgruppe FOR 1234 iBeetle: Functional Genomics of Insect Development and Metamorphosis).
